Introduction
Example 5.2: Cobb Douglas production function (Cobb and Douglas, 1928)
- Cobb Douglas production:
- Apply a logarithmic transformation
- The values of
OLS estimation: Model Setup
- Define the column vectors
OLS estimation: Model Setup
-
LRM in matrix form
-
-
Ordinary Least Squares (OLS) criterion
-
OLS estimator of
OLS estimation
-
We can interpret the regression coefficient as the marginal effect,
- which is represented by
- which is represented by
-
An increase of one unit in
- the expected value of
- while keeping all other factors constant.
- the expected value of
-
The relationship between a dependent variable and independent variables doesn't always have to be linear.
OLS estimation: Properties of OLS estimator
- The OLS estimator remains unbiased.
- Variance-covariance matrix has the form of
OLS Estimation
Use the dataset (data_incomeedu.dta) provided in the supplementary material to illustrate OLS estimation result in Python.
------------------------------------------------
variable name type variable label
------------------------------------------------
pid int pseudo ID (respondent)
village_pid int pseudo ID (village)
educ int years of education
income float income in 2020 (RMB)
------------------------------------------------
OLS Estimation
import pandas as pd
import numpy as np
# 对应目录下从data_wageedu.dta中读取数据
data = pd.pandas.read_stata("dependency/data_incomeedu.dta")
x = data.loc[:,"educ"].to_numpy(dtype='float',
na_value=np.nan).reshape(-1,1)
y = data.loc[:,"income"].to_numpy(dtype='float',
na_value=np.nan).reshape(-1,1)
y = np.log(y)
X = np.append(np.ones((y.size,1)), x, axis=1)
OLS Estimation: Efficiency
import time
# 方法一:循环计算
def forloopols(x, y):
x_bar = x.mean()
y_bar = y.mean()
denominator = 0
numerator = 0
for i in range(y.size):
denominator += (x[i]-x_bar)**2
numerator += (x[i]-x_bar)*(y[i]-y_bar)
return numerator/denominator
# 方法二:矩阵计算
def matols(x, y):
X = np.append(np.ones((y.size,1)),x,axis=1)
return np.matmul(np.matmul(np.linalg.inv(np.matmul(X.T,X)), X.T), y)[1]
startTime = time.time() # 计时开始
for k in range(1000):
slope_v1, = forloopols(x, y)
endTime = time.time()-startTime # 计时结束
startTime = time.time()
for k in range(1000):
slope_v2, = matols(x, y)
faster_rate = endTime/(time.time()-startTime)
print("beta2 is estimated to be %5.3f by for-loop"
% (slope_v1))
print("beta2 is estimated to be %5.3f by matrix computation"
% (slope_v2))
print("Matrix computation is ", faster_rate," times faster.")
beta2 is estimated to be 0.039 by for-loop
beta2 is estimated to be 0.039 by matrix computation
Matrix computation is 34.75705486770159 times faster.
# 使用 np.dot()命令进行计算
print("By np.dot():",
np.dot(np.dot(np.linalg.inv(np.dot(X.T,X)), X.T), y)[1])
# 使用 np.matmul()命令进行计算
print("By np.matmul():",
np.matmul(np.matmul(np.linalg.inv(np.matmul(X.T,X)), X.T),
y)[1])
# 使用 @ 命令进行计算
print("By @:",(np.linalg.inv(X.T @ X)@ X.T @ y )[1])
By np.dot(): [0.03915481]
By np.matmul(): [0.03915481]
By @: [0.03915481]
def timer_ols(method, rep_time=500000):
startTime = time.time() # 计时开始
if method == "dot": # if条件为DOT的方法
for ii in range(rep_time):
np.dot(np.dot(np.linalg.inv(np.dot(X.T,X)), X.T), y)
print("np.dot() takes %6.5f seconds"
% (time.time()-startTime) ) # 输出时长
elif method == "matmul":# if条件为MATMUL的方法
for ii in range(rep_time):
np.matmul(np.matmul(np.linalg.inv(np.matmul(X.T,X)),
X.T), y)
print("np.matmul() takes %6.5f seconds"
% (time.time()-startTime)) # 输出时长
elif method == "at": # if条件为@的方法
for ii in range(rep_time):
(np.linalg.inv(X.T @ X) @ X.T) @ y
print("@ takes %6.5f seconds"
% (time.time()-startTime)) # 输出时长
# 执行
timer_ols("dot")
timer_ols("matmul")
timer_ols("at")
np.dot() takes 13.22670 seconds
np.matmul() takes 10.44645 seconds
@ takes 10.75469 seconds
OLS Estimation
- To estimate
np.linalg.lstsq(X, y, rcond=None)[0]
array([[9.99572763],
[0.03915481]])
- We can also estimate
- One can apply the
statsmodels.OLSroutine. add_constant()function could add the constant term directly into the covariates matrix.
OLS Estimation
import statsmodels.api as sm
X = sm.add_constant(x)
# 采用回归方程进行数据输入并进行拟合以及估计
result_ols = sm.OLS(y, X).fit()
print(result_ols.summary())
OLS Regression Results
==============================================================================
Dep. Variable: y R-squared: 0.041
Model: OLS Adj. R-squared: 0.032
Method: Least Squares F-statistic: 4.778
Date: Wed, 26 Jul 2023 Prob (F-statistic): 0.0309
Time: 14:39:54 Log-Likelihood: -96.436
No. Observations: 115 AIC: 196.9
Df Residuals: 113 BIC: 202.4
Df Model: 1
Covariance Type: nonrobust
==============================================================================
coef std err t P>|t| [0.025 0.975]
------------------------------------------------------------------------------
const 9.9957 0.159 62.705 0.000 9.680 10.312
x1 0.0392 0.018 2.186 0.031 0.004 0.075
==============================================================================
Omnibus: 94.673 Durbin-Watson: 1.053
Prob(Omnibus): 0.000 Jarque-Bera (JB): 1055.552
Skew: 2.642 Prob(JB): 6.16e-230
Kurtosis: 16.870 Cond. No. 27.2
==============================================================================
Notes:
[1] Standard Errors assume that the covariance matrix of the errors is correctly specified.
OLS Estimation
import matplotlib.pyplot as plt
beta = np.linalg.inv((X.T @ X))@ X.T @ y
plt.figure(figsize=(8,5))
plt.plot(x, y, 'ok', label='Original data', markersize=10)
plt.plot(x, beta[0] + beta[1]*x, 'gray', label='Fitted line')
plt.title("Figure 5.1: The OLS Fitted Line and the Original Data",
fontsize=14)
plt.xlabel(r"$x$", fontsize=12)
plt.ylabel(r"$y$", fontsize=12)
plt.legend(fontsize=12)
plt.show()
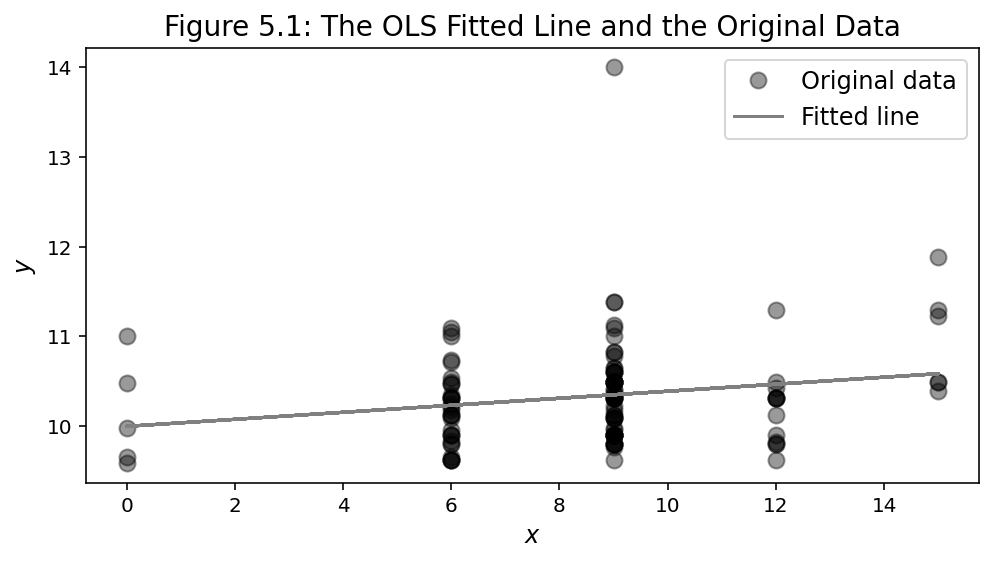
OLS Estimation
- As
-
Therefore,
-
One could compute the variance directly in Python based on this formula.
OLS Estimation
N = y.size
beta = np.dot(np.dot(np.linalg.inv(np.dot(X.T,X)), X.T), y)
e = y-np.dot(X,beta)
# 计算sigma平方的估计量
sigma2 = (1/(N-2)) * np.inner(e.T,e.T)
print("Estimate of sigma2:",sigma2[0])
# 计算OLS估计量方差矩阵
var = np.linalg.inv(np.dot(X.T,X))*sigma2
print("Variance-covariance matrix \n", var)
# 计算OLS估计量标准误
se = np.sqrt(np.diag(var))
print("Standard errors of beta0 is %6.3f and of beta1 is %6.3f"
% (se[0],se[1]))
OLS Estimation
Estimate of sigma2: [0.31879951]
Variance-covariance matrix
[[ 0.02541155 -0.00269516]
[-0.00269516 0.00032085]]
Standard errors of beta0 is 0.159 and of beta1 is 0.018
OLS Estimation
- Assessing goodness-of-fit in regression analysis.
- coefficient of determination, denoted as
- TSS (total sum of squares), RSS (residual sum of squares), and ESS (error sum of squares) can be written in the following scalar notations
- The adjusted
OLS Estimation
# 定义M0矩阵
n = y.size
k = X.shape[1]
M0 = np.eye(n) - np.ones((n,n))/n
# 计算RSS,TSS与ESS
RSS = e.T@e
TSS = y.T@M0@y
# 计算R2与adj. R2
R2 = 1 - RSS/TSS
adjR2 = 1- (RSS/(n-k))/(TSS/(n-1))
print("R-squared: %5.4f" % (R2))
print("Adj. R-squared: %5.4f" % (adjR2))
R-squared: 0.0406
Adj. R-squared: 0.0321
OLS Estimation
To verify this equation, we can utilize the following code:
print("Adj. R-squared: %5.4f" % ((1- (n-1)*(1-R2)/(n-k))))
Adj. R-squared: 0.0321
Inferences
- Conducting a hypothesis test on a single restriction:
Consider again
- We have a conjecture that people with higher educational levels would earn more, so we anticipate rejecting the null hypothesis.
Inferences
print("t-stat of beta1: %4.3f"
% ((beta.reshape(-1)/se)[1]) )
t-stat of beta1: 2.186
from scipy.stats import t
cv = t.ppf(0.975, df=N-2) # 临界值
lower_bounds = beta.reshape(-1) - se*cv
upper_bounds = beta.reshape(-1) + se*cv
print("95 CIs of beta1: [%5.3f, %5.3f]" % (lower_bounds[1],upper_bounds[1]))
95 CIs of beta1: [0.004, 0.075]
Inferences
- As we do not observe the error term
from scipy.stats import norm
# 计算S_x与S_ex
Sx = (X.T@X)/n
Q = np.diag(e.reshape(-1))**2
Sex = (X.T@Q@X)/n
# 计算渐近方差
Avar = np.linalg.inv(Sx)@Sex@np.linalg.inv(Sx)/n
Ase = np.sqrt(np.diag(Avar))
# 计算z统计量
zstat = beta.reshape(-1)/Ase
print("z-stat beta1 is %4.3f" % (zstat[1]) )
# 渐近置信区间
cv = norm.ppf(0.975)
lower_bounds = beta.reshape(-1) - Ase*cv
upper_bounds = beta.reshape(-1) + Ase*cv
print("95 asy. CIs of beta1 is [%5.3f,%5.3f]"
% (lower_bounds[1],upper_bounds[1]))
z-stat beta1 is 2.223
95 asy. CIs of beta1 is [0.005,0.074]
Inferences
-
- The restriction can be expressed as
R = [[1, -1]]
print(result_ols.f_test(R))
<F test: F=3185.238490556453, p=1.2527722221299107e-84, df_denom=113, df_num=1>
Residual Analysis
- The residual analysis allows us to check the adequacy of the model and detect outliers.
- It is common to study the distribution of residuals by plotting the histogram.
- Standardize residuals as
- is the estimated standard error for
- is the estimated standard error for
- Then reconcile the plots
Residual Analysis
import seaborn as sns
# 标准化残差
sigma2_e = np.sqrt(np.sum(e**2)/(N-2))
r = e/sigma2_e
reference = np.random.normal(size=200000)
# 作图
plt.figure(figsize=(8,5))
sns.kdeplot(reference, color='k',
label=r"$N(0,1)$ PDF", linestyle="--")
sns.histplot(r, kde=True, linewidth=0, color='gray',
kde_kws={'cut':3}, stat='density',
label=r"Standardized residuals $r_i$")
plt.title("Figure 5.2: Visualization the Standardized Residuals",
fontsize=14)
plt.ylabel("density", fontsize=12)
plt.xlabel(r"$r_i$", fontsize=12)
plt.legend(loc="upper right", fontsize=12)
plt.show()
<><>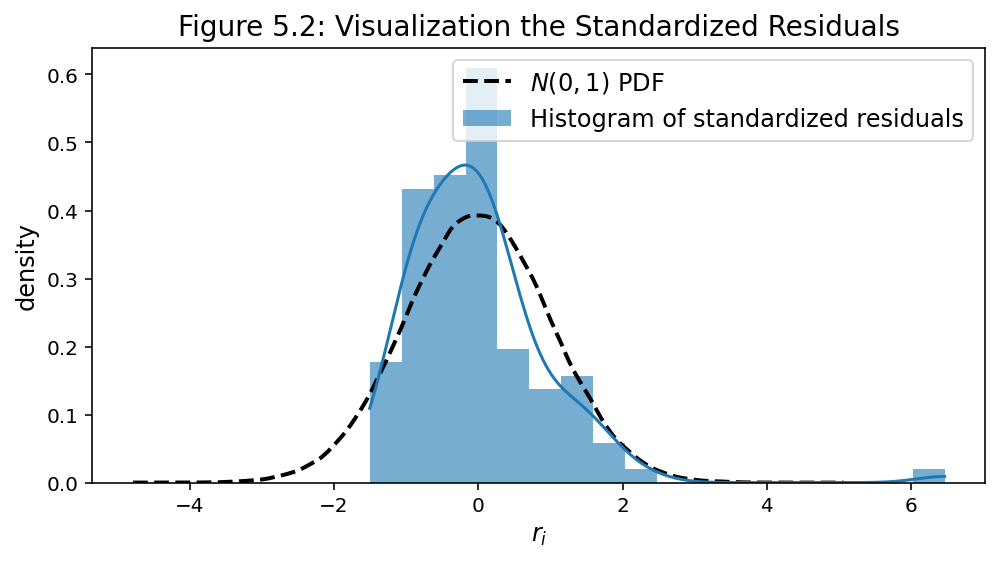
Residual Analysis: Test for the Normality
from scipy.stats import normaltest
k2, p = normaltest(r)
print("K2 statistic: %5.4f \np-value: %5.4f" % (k2,p))
K2 statistic: 94.6726
p-value: 0.0000
Residual Analysis: Test for Heteroskedasticity
-
A range of testing statistics to detect heteroscedasticity in the residuals.
-
Examples Breusch-Pagan (BP) test (Breusch and Pagan, 1979) and the White test (White, 1980).
-
Residual Analysis: Test for Heteroskedasticity
- Test for heteroskedasticity means that all slope coefficients are equal to zero.
- This involves testing if
- The intercept
- The intercept
- As we cannot observe
Residual Analysis: Test for Heteroskedasticity
from statsmodels.stats.diagnostic import het_breuschpagan, het_white
model = sm.OLS(y, X).fit()
bp_result = het_breuschpagan(model.resid, X)
white_result = het_white(model.resid, X)
print('BP test p-value: '+str(bp_result[3]))
print('White test p-value: '+str(white_result[3]))
BP test p-value: 0.6762218491291369
White test p-value: 0.9152386628311834
Robust Standard Errors: White Standard Errors
- The errors are most likely heteroskedastic.
- One common technique is to use the White standard errors (White, 1980).
- These are also known as
- heteroskedasticity-robust standard errors,
- or Eicker-Huber-White standard errors
Consider the formula:
We code in Python next.
Robust Standard Errors
# 计算Omega
omega_hat = (e@e.T)*n/(n-k)
# 计算稳健方差矩阵
XXinv = np.linalg.inv(X.T@X)
var = XXinv@X.T@np.diag(np.diag(omega_hat))@X@XXinv
print("White variance-covariance matrix \n", var)
# 计算稳健标准误
se = np.sqrt(np.diag(var))
print("White standard errors of beta0 is %6.5f and of beta1 is %6.5f"
% (se[0],se[1]))
White variance-covariance matrix
[[ 0.02255978 -0.00250354]
[-0.00250354 0.00031564]]
White standard errors of beta0 is 0.15020 and of beta1 is 0.01777
Robust Standard Errors: Clustered Standard Errors
- clustered standard errors are widely used in a variety of applied econometric settings.
- The sample consists of survey responses from 110 individuals across 8 randomly selected villages in Guangdong Province.
- 8 villages can be situated in different areas of Guangdong. Therefore, it is likely that respondents from more developed villages have a higher income compared to those from less developed ones.
Call Stata from Python
# 配置Python-Stata交互
import os
os.chdir('/Applications/Stata/utilities')
from pystata import config
config.init('mp')
# 将数据传输至Stata
from pystata import stata
stata.pdataframe_to_data(data, force=True)
# 回归并使用聚类标准误
stata.run(
'''gen lincome = ln(income)
regress lincome educ, cluster(village_pid)
''')
Linear regression Number of obs = 115
F(1, 7) = 9.80
Prob > F = 0.0166
R-squared = 0.0406
Root MSE = .56462
(Std. err. adjusted for 8 clusters in village_pid)
---------------------------------------------------------------------
| Robust
lincome | Coefficient std. err. t P>|t| [95% conf. interval]
--------+------------------------------------------------------------
educ | .0391548 .0125084 3.13 0.017 .0095772 .0687324
_cons | 9.995728 .1576285 63.41 0.000 9.622995 10.36846
---------------------------------------------------------------------
Robust Standard Errors: Clustered Standard Errors
- For the LRM, the cluster standard errors can be expressed as
Robust Standard Errors: Clustered Standard Errors
# 从数据中获取cluster变量
cluster = data.loc[:,"village_pid"].to_numpy(dtype='float',
na_value=np.nan).reshape(-1,1)
G = len(np.unique(cluster))
# 计算聚类方差的Omega
omega_hat = (e@e.T)*((G)/(G-1))*((n-1)/(n-k))
omega_hat_c = np.copy(omega_hat)
for i in range(n):
for j in range(n):
if i>j:
if cluster[i]!=cluster[j]:
omega_hat_c[i,j] = 0
omega_hat_c[j,i] = 0
# 计算聚类方差矩阵
XXinv = np.linalg.inv(X.T@X)
var = XXinv@X.T@omega_hat_c@X@XXinv
print("Clustered variance-covariance matrix \n", var)
# 计算聚类标准误
se = np.sqrt(np.diag(var))
print("Clustered standard errors of beta0 is %6.5f and of beta1 is %6.5f"
% (se[0],se[1]))
Clustered variance-covariance matrix
[[ 0.02484674 -0.00062565]
[-0.00062565 0.00015646]]
Clustered standard errors of beta0 is 0.15763 and of beta1 is 0.01251
Endogeneity
- When studying the return of education problems, the economic literature (Mroz, 1987; Card, 1993) usually employ a linear model to estimate the impact of years of schooling (
- The OLS estimator is biased. This bias is known as omitted variable bias.
- Omitted variables that are correlated with dependent and independent variables
- Independent variables is measured with error
- Dependent and independent variables that are simultaneously determined.
Endogeneity: IV Estimations
- To conduct IV estimations, IV variables meet two conditions:
- IV exogeneity: IV variables are uncorrelated with the error term.
- IV relevance : IV variables should be partially and sufficiently strongly correlated with the endogeneous variable once the other independent variables are controlled for.
Endogeneity: Just-identified Case
-
We cannot assume that the
-
IV conditions are represented as:
- Since
Endogeneity: Just-identified Case
- The IV estimator of
- Define
Endogeneity: Just-identified Case
- Note that this estimator is valid under homoskedasticity. We will discuss heteroskedastic case later.
Endogeneity: Just-identified Case
import wooldridge
import pandas as pd
import statsmodels.api as sm
import statsmodels.formula.api as smf
# 读取数据包当中的MORZ原始数据
reduc_women = wooldridge.data('mroz')
# 去掉数据中工资为空的观测值
datas = reduc_women.dropna(subset=['lwage'])
x=datas[['exper','expersq','educ']]
y=datas.loc[:,'lwage']
X = sm.add_constant(x)
# OLS估计法(不处理内生性问题)
result_ols = sm.OLS(y, X).fit()
print(result_ols.summary2())
Results: Ordinary least squares
=================================================================
Model: OLS Adj. R-squared: 0.151
Dependent Variable: lwage AIC: 871.1979
Date: 2023-11-15 17:58 BIC: 887.4344
No. Observations: 428 Log-Likelihood: -431.60
Df Model: 3 F-statistic: 26.29
Df Residuals: 424 Prob (F-statistic): 1.30e-15
R-squared: 0.157 Scale: 0.44412
-------------------------------------------------------------------
Coef. Std.Err. t P>|t| [0.025 0.975]
-------------------------------------------------------------------
const -0.5220 0.1986 -2.6282 0.0089 -0.9125 -0.1316
exper 0.0416 0.0132 3.1549 0.0017 0.0157 0.0675
expersq -0.0008 0.0004 -2.0628 0.0397 -0.0016 -0.0000
educ 0.1075 0.0141 7.5983 0.0000 0.0797 0.1353
-----------------------------------------------------------------
Omnibus: 77.792 Durbin-Watson: 1.961
Prob(Omnibus): 0.000 Jarque-Bera (JB): 300.917
Skew: -0.753 Prob(JB): 0.000
Kurtosis: 6.822 Condition No.: 2212
=================================================================
* The condition number is large (2e+03). This might indicate
strong multicollinearity or other numerical problems.
Endogeneity: IV Estimations
y = datas[['lwage']].to_numpy()
X = sm.add_constant(datas[['exper','expersq','educ']]).to_numpy()
Z = sm.add_constant(datas[['exper','expersq',
'fatheduc']]).to_numpy()
# IV点估计
beta_IV = np.linalg.inv(Z.T@X)@Z.T@y
# 方差估计
e = y-X@beta_IV
# 计算sigma平方的估计量
sigma2 = (1/(y.size)) * np.inner(e.T,e.T)
se_IV = np.sqrt(np.diag(np.linalg.inv(Z.T@X)@Z.T
@Z@np.linalg.inv(X.T@Z)*sigma2))
Endogeneity: IV Estimations
# 汇报结果
print(" Parameter Std. Err.")
print("Intercept %8.4f, %8.4f"
% (beta_IV[0], se_IV[0]) )
print("exper %8.4f, %8.4f"
% (beta_IV[1], se_IV[1]) )
print("expersq %8.4f, %8.4f"
% (beta_IV[2], se_IV[2]) )
print("educ %8.4f, %8.4f"
% (beta_IV[3], se_IV[3]) )
Parameter Std. Err.
Intercept -0.0611, 0.4344
exper 0.0437, 0.0133
expersq -0.0009, 0.0004
educ 0.0702, 0.0343
Endogeneity: IV Estimations
from linearmodels.iv import IV2SLS
reg_2sls=IV2SLS.from_formula(
formula='lwage~1+exper+expersq+[educ~fatheduc]',
data= datas)
print(reg_2sls.fit(cov_type='unadjusted'))
IV-2SLS Estimation Summary
===================================================================
Dep. Variable: lwage R-squared: 0.1430
Estimator: IV-2SLS Adj. R-squared: 0.1370
No. Observations: 428 F-statistic: 25.176
Date: Wed, Nov 15 2023 P-value (F-stat) 0.0000
Time: 17:57:30 Distribution: chi2(3)
Cov. Estimator: unadjusted
Parameter Estimates
===================================================================
Parameter Std. Err. T-stat P-value Lower CI Upper CI
-------------------------------------------------------------------
Intercept -0.0611 0.4344 -0.1407 0.8881 -0.9125 0.7903
exper 0.0437 0.0133 3.2744 0.0011 0.0175 0.0698
expersq -0.0009 0.0004 -2.2107 0.0271 -0.0017 -0.0001
educ 0.0702 0.0343 2.0485 0.0405 0.0030 0.1374
===================================================================
Endogenous: educ
Instruments: fatheduc
Unadjusted Covariance (Homoskedastic)
Debiased: False
Endogeneity: 2SLS Estimation
- the projection matrix
- We use
Endogeneity: 2SLS Estimation
The two-step procedure can be described as:
-
Obtain
-
Use
We can just estimate 2SLS estimators in one step by using
Endogeneity: 2SLS Estimation
Z = sm.add_constant(datas[['exper','expersq','fatheduc',
'motheduc']]).to_numpy()
# 2SLS点估计
Pz = Z@np.linalg.inv(Z.T@Z)@Z.T
Xhat = Pz@X
beta_2SLS = np.linalg.inv(Xhat.T@X)@Xhat.T@y
Endogeneity: 2SLS Estimation
- The variance (or standard errors) produced by the two-step procedure are incorrect and tend to be smaller than the actual ones.
- The correct variance (under homoskedastric assumption)
-
-
The heteroskedasticity-robust (White) standard errors is
Endogeneity: 2SLS Estimation
# 方差估计
e = y-X@beta_2SLS
# 计算sigma平方的估计量
diagomega = np.diag(np.diag(e@e.T))
se_2SLS = np.sqrt(np.diag(np.linalg.inv(Xhat.T@Xhat)
@(Xhat.T@diagomega@Xhat)
@np.linalg.inv(Xhat.T@Xhat) ))
print(" Parameter Robust Std. Err.")
print("Intercept %8.4f, %8.4f"
% (beta_2SLS[0], se_2SLS[0]) )
print("exper %8.4f, %8.4f"
% (beta_2SLS[1], se_2SLS[1]) )
print("expersq %8.4f, %8.4f"
% (beta_2SLS[2], se_2SLS[2]) )
print("educ %8.4f, %8.4f"
% (beta_2SLS[3], se_2SLS[3]) )
Parameter Robust Std. Err.
Intercept 0.0481, 0.4278
exper 0.0442, 0.0155
expersq -0.0009, 0.0004
educ 0.0614, 0.0332
from linearmodels.iv import IV2SLS
reg_2sls=IV2SLS.from_formula(
formula='lwage~1+exper+expersq+[educ~motheduc+fatheduc]',
data= datas)
print(reg_2sls.fit())
IV-2SLS Estimation Summary
===================================================================
Dep. Variable: lwage R-squared: 0.1357
Estimator: IV-2SLS Adj. R-squared: 0.1296
No. Observations: 428 F-statistic: 18.611
Date: Wed, Nov 15 2023 P-value (F-stat) 0.0003
Time: 17:58:25 Distribution: chi2(3)
Cov. Estimator: robust
Parameter Estimates
===================================================================
Parameter Std. Err. T-stat P-value Lower CI Upper CI
-------------------------------------------------------------------
Intercept 0.0481 0.4278 0.1124 0.9105 -0.7903 0.8865
exper 0.0442 0.0155 2.8546 0.0043 0.0138 0.0745
expersq -0.0009 0.0004 -2.1001 0.0357 -0.0017 -0.0001
educ 0.0614 0.0332 1.8503 0.0643 -0.0036 0.1264
===================================================================
Endogenous: educ
Instruments: fatheduc, motheduc
Unadjusted Covariance (Heteroskedastic)
Debiased: False
Endogeneity: Testing for IV Validity
- The first-step estimation of the 2SLS method can be used to test for the IV relevance.
- Conduct an
#第一阶段回归
reg_1stage=smf.ols(formula='educ~exper+expersq+fatheduc',
data=datas)
results_1stage=reg_1stage.fit()
print(results_1stage.summary2())
Results: Ordinary least squares
==================================================================
Model: OLS Adj. R-squared: 0.170
Dependent Variable: educ AIC: 1846.5075
Date: 2023-11-15 19:53 BIC: 1862.7440
No. Observations: 428 Log-Likelihood: -919.25
Df Model: 3 F-statistic: 30.09
Df Residuals: 424 Prob (F-statistic): 1.18e-17
R-squared: 0.176 Scale: 4.3366
-------------------------------------------------------------------
Coef. Std.Err. t P>|t| [0.025 0.975]
-------------------------------------------------------------------
Intercept 9.8870 0.3956 24.9920 0.0000 9.1094 10.6646
exper 0.0468 0.0411 1.1391 0.2553 -0.0340 0.1276
expersq -0.0012 0.0012 -0.9364 0.3496 -0.0036 0.0013
fatheduc 0.2705 0.0289 9.3670 0.0000 0.2137 0.3273
------------------------------------------------------------------
Omnibus: 10.235 Durbin-Watson: 1.919
Prob(Omnibus): 0.006 Jarque-Bera (JB): 17.236
Skew: -0.103 Prob(JB): 0.000
Kurtosis: 3.961 Condition No.: 1411
==================================================================
* The condition number is large (1e+03). This might indicate
strong multicollinearity or other numerical problems.
Endogeneity: Testing for IV Validity
- In the case of having more instrumental variables than endogenous variables,
- which is known as the over-identified case,
- We can use the over-identification test to confirm this requirement.
- The intuition:
- if we have two valid instruments, such as
- we can compute two separate estimators.
- if we have two valid instruments, such as
- If these estimators are significantly different from each other,
- it indicates that at least one of the instrumental variables is invalid.
Endogeneity: Testing for IV Validity
from scipy.stats import chi2
# 1. 2SLS回归,并计算残差项
reg_2sls=IV2SLS.from_formula(
formula='lwage~1+exper+expersq+[educ~fatheduc+motheduc]',
data=datas)
results_2sls=reg_2sls.fit(cov_type='unadjusted', debiased=True)
# 2. 残差对工具变量与外生变量回归
datas['resid_2sls']=results_2sls.resids
# 3.1 检验OR所需要的回归构建
reg_or=smf.ols(formula='resid_2sls~ 1+exper+expersq+motheduc+fatheduc', data=datas)
results_or=reg_or.fit()
Endogeneity: Testing for IV Validity
# 3.2 统计检验得出结论
R2 = results_or.rsquared
n = results_or.nobs
q = 2-1
teststt=n*R2
pval=1-chi2.cdf(teststt,q)
print(f'Overidentification test: {teststt}')
print(f'p-value: {pval}')
print(f'sample size: {n}')
Overidentification test: 0.3780713419639192
p-value: 0.5386372330714363
sample size: 428.0
Endogeneity: Testing for Endogeneity
# 第一阶段回归
reg_1stage=smf.ols(formula='educ~exper+expersq+motheduc+fatheduc',
data=datas)
results_1stage=reg_1stage.fit()
# 从计算结果中提取残差项
datas['resid']=results_1stage.resid
# 将残差项加入OLS模型
reg_endo=smf.ols(formula='lwage~resid+educ+exper+expersq',data=datas)
results_endo=reg_endo.fit()
print(results_endo.summary2())
Results: Ordinary least squares
=================================================================
Model: OLS Adj. R-squared: 0.154
Dependent Variable: lwage AIC: 870.3816
Date: 2023-11-15 20:15 BIC: 890.6772
No. Observations: 428 Log-Likelihood: -430.19
Df Model: 4 F-statistic: 20.50
Df Residuals: 423 Prob (F-statistic): 1.89e-15
R-squared: 0.162 Scale: 0.44225
------------------------------------------------------------------
Coef. Std.Err. t P>|t| [0.025 0.975]
------------------------------------------------------------------
Intercept 0.0481 0.3946 0.1219 0.9030 -0.7275 0.8237
resid 0.0582 0.0348 1.6711 0.0954 -0.0103 0.1266
educ 0.0614 0.0310 1.9815 0.0482 0.0005 0.1223
exper 0.0442 0.0132 3.3363 0.0009 0.0181 0.0702
expersq -0.0009 0.0004 -2.2706 0.0237 -0.0017 -0.0001
-----------------------------------------------------------------
Omnibus: 74.968 Durbin-Watson: 1.931
Prob(Omnibus): 0.000 Jarque-Bera (JB): 278.059
Skew: -0.736 Prob(JB): 0.000
Kurtosis: 6.664 Condition No.: 4419
=================================================================